Thomas Schmidt Lab - Single Molecule Microscopy
Intrigued by the way cells autonomously regulate their fate, we strive to understand and visualize cellular processes as the basis of Cell Signaling.
Although extensive knowledge exists about the molecular players, their structure, and their respective interactions, in many cellular signaling pathways the physical and mechanistic properties underlying signaling processes still need to be fully understood. Those cellular processes are ultimately diffusion controlled, hence largely unsynchronized, making them less accessible to typical biochemical bulk methods used so far.
Single-Molecule Microscopy
We set out to apply and develop in vivo Single-Molecule Microscopy methods in order to follow the molecular players one-by-one and in real time, thereby obtaining direct information on the physical and mechanistic processes underlying cellular signaling processes. Whereas studies of in vivo systems set the link to biological relevance, complementary in vitro studies do allow us to rigorously test the molecular/mechanistic models which we infer from the in vivo results.
Cellular signaling
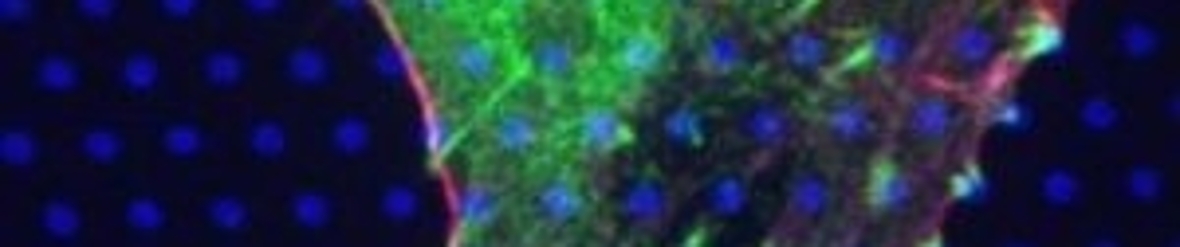
In our efforts we concentrate mainly on the early events in cellular signaling, the moment e.g. a ligand binds to a receptor. Most of such processes take place on the Plasma Membrane of the cell. In relation to signaling processes the two-dimensional environment of the plasma membrane, and its potential nanostructuring, is believed to play a major role in the controlled and robust functioning of signaling pathways. Our emphasis is on unraveling directional sensing as governed by G-Protein Coupled Receptors, and on the initial events that leads to cellular Mechanosensing.