Research project
Structure and regulation in photosynthesis
Plants, algae and cyanobacteria have the amazing capacity to perform delicate photophysical and photochemical processes of light capture, excitation transfer, charge separation and catalysis in fluctuating living environments. As a consequence, photosynthetic proteins and membranes form dynamic architectures. We study their flexible structures and performances to understand the mechanisms underlying dynamic photosynthetic energy conversion. Ultimately, this information may be used to re-design species that convert more light into biomass or to design solar-energy converting artificial constructs based on biological design principles.
- Contact
- Anjali Pandit
Living photosynthetic organisms are capable to activate processes by light and to convert light energy into the storage of chemical bonds. We want to know how these processes are carried out by chromophores, tuned by a protein scaffold and controlled by a responsive membrane. NMR spectroscopy is the method of choice, because of its ability to detect structure and dynamics with atomistic resolution and its application to samples in physiological conditions.
In solution NMR, linewidths increase as the molecules get larger and anisotropic interactions that influence the behavior of nuclear spins are not average to zero by brownian motions. Most of the time, our systems of interest are membrane proteins, biological membranes or even entire cells, which are considerably large. Therefore we apply solid-state NMR (ssNMR) spectroscopy. In ssNMR, samples are concentrated into small rotors and spun fast under a magic angle with respect to the static magnetic field to average anisotropic interactions. Relaxation measurements and dynamic spectral editing are applied as specific tools to detect dynamic motions of proteins, ligands and (other) cell constituents.

Molecular photo switches
Photosynthesis is tightly regulated via a feedback network that controls the activity of the light-harvesting antenna and prevents photodamage. The key players in this process are membrane proteins that act as molecular switches and respond to changes in light or pH. We study their functional mechanisms by combination of NMR and optical spectroscopy. We have shown that the micro-environment has considerable influence on the conformational dynamics of membrane proteins and use reconstitution in detergent micelles, liposomes or tailored lipid nanodiscs to address the role of a membrane environment.
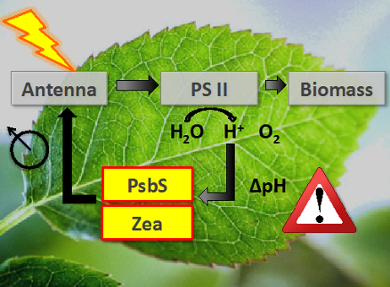
Towards in-cell NMR
In-cell solid-state NMR is a rapid emerging field of research that is confronted with the major challenge of identifying atomistic signals from targeted biomolecules against the background of all other cell components. We use the green alga Chlamydomonas reinhardtii as a model organism to explore the exciting opportunities of in-situ and in-cell NMR.
Using NMR dynamic spectral editing, we have demonstrated that we can detect the conformational dynamics of a target protein in native photosynthetic membranes and probe molecular dynamics inside chloroplast membranes of intact cells. We collaborate with plant biochemists and biophysicists to understand the role of membrane dynamics in the regulation of photosynthesis.

Artificial proteins for solar-energy conversion
In natural photosynthesis, proteins have a crucial role in optimizing excitation and electron transfer by positioning of chromophores and providing a scaffold for catalytic reactions to take place. In collaboration with inorganic chemists in the LIC (Bonnet lab), we work towards the design of semi-artificial metalloproteins for photocatalytic water oxidation. Modified proteins may offer a protected molecular environment where artificial water oxidation catalysts and photosensitizers can be organized in a controlled fashion, to perform efficient (photo)catalytic water oxidation while minimizing the rate of decomposition. In collaboration with theoretical chemists in the LIC (Dr. Francesco Buda) we are interested in protein-ligand interactions in retinal-binding proteins, for rational design of novel photo proteins with controllable optical and functional properties.
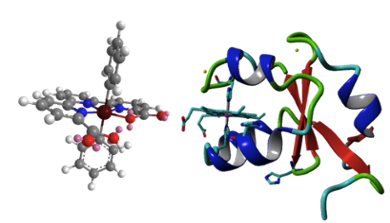
Key publications
- Azadi‑Chegeni F., Schiphorst C. & Pandit A. (2018), In vivo NMR as a tool for probing molecular structure and dynamics in intact Chlamydomonas reinhardtii cells, Photosynthesis Research 135(1-3): 227-237.
- Crisafi E. & Pandit A. (2017), Disentangling protein and lipid interactions that control a molecular switch in photosynthetic light harvesting, BIOCHIMICA ET BIOPHYSICA ACTA-BIOMEMBRANES 1859: 40-47.
- Krishnan M., Moolenaar G.F., Sai Sankar Gupta K.B., Goosen N. & Pandit A. (2017), Large-scale in vitro production, refolding and dimerization of PsbS in different microenvironments, Scientific Reports 7: 15200.
- Azadi Chegeni F., Perin, G., Sai Sankar Gupta K.B., Simionato D., Morosinotto T. & Pandit A. (2016), Protein and lipid dynamics in photosynthetic thylakoid membranes investigated by in-situ solid-state NMR, BIOCHIMICA ET BIOPHYSICA ACTA-BIOENERGETICS1857(12): 1849–1859.
- Mathes T., Heilmann M.a, Pandit A., Zhu J., Ravensbergen J., Kloz M., Fu Y., Smith B.O., Christie J.M., Jenkins G.I. & Kennis J.T.M. (2015), Proton-Coupled Electron Transfer Constitutes the Photoactivation Mechanism of the Plant Photoreceptor UVR8, Journal of the American Chemical Society 137(25): 8113-8120.
- Pandit A., Ocakoglu K., Buda F., Marle Th. van, Holzwarth A.R. & Groot H.J.M. de (2013), Structure determination of a bio-inspired self-assembled light-harvesting antenna by solid-state NMR and molecular modeling, 117: 11292-11298.
